
RNA-seq and pathway analysis
RNA-sequencing (RNA-seq) has evolved into an indispensable tool for analysing gene expression levels, offering profound insights into the cellular transcriptome. It is an extremely valuable technique for comparing conditions such as treatment versus control or knockout versus control.
Our approach to analysing your RNA-seq data is centred around your specific interests and needs:
Quality checks: thorough quality checks to ensure the reliability of the dataset
Differential expression analysis:
Statistical comparisons between two sample groups unveil crucial information about fold changes and statistical significance for each identified transcript.
Visual representation through heat maps, box plots, volcano plots, and other informative graphics.
Pathway analysis:
Placing the identified differential expression levels within a broader biological context.
Unraveling the changes in biological processes to provide a comprehensive understanding.
You can also check our publication list to see more examples.
RNA-seq, an indispensable tool
Our commitment extends beyond the analysis process. If you’re planning a gene expression experiment and have queries about the wet lab aspects, don’t hesitate to contact us for pre-experiment consultancy advice! We can guide you on crucial decisions, such as selecting the appropriate library preparation kit, determining whether single-end or paired-end sequencing is suitable (tip: check our video blogs) and advising on the choice between polyA enrichment or rRNA depletion. Your experiment's success is our priority, and we're here to assist you at every stage.
Heatmaps are often a good way to show changes between samples (Lambros et al. 2023, IJMS)

Venn diagram showing differentially expressed genes and similarities between groups

Pathway differences
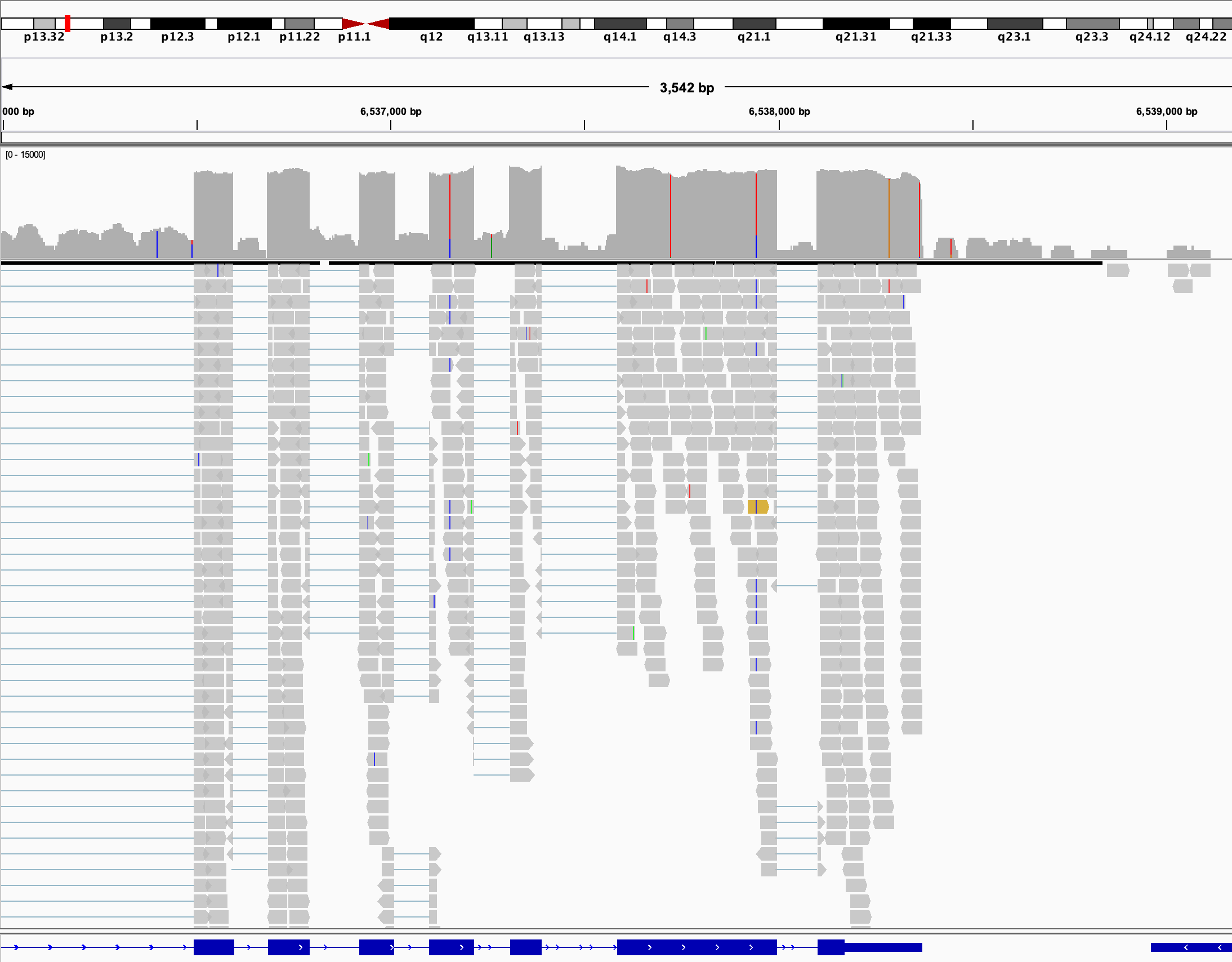
Screenshot of a gene in a genome browser, clearly showing the different introns and exons.
Volcano Plot showing differentially expressed genes with colour indication of the relevant genes for this study
Example of a coverage analysis on mitochondrial RNA (Van Haute et al. 2023, Nature Communications)




