Metagenomic sequencing
Metagenomics is a groundbreaking field that involves the direct study of genetic material extracted from environmental samples like soil, water, or air rather than from cultivated organisms. Metagenomics enables the exploration of entire microbial communities within ecosystems, encompassing bacteria, archaea, viruses, and eukaryotes. The goal is to unveil these organisms' functional and structural diversity. Metagenomics finds application across diverse disciplines such as ecology, medicine, biotechnology, and agriculture.
Several techniques are employed in metagenomics:
Shotgun Sequencing:
It involves randomly fragmenting DNA and sequencing the resulting fragments.
Targeted Sequencing:
Focuses on specific genome regions or organisms within the community.
16S/18S/ITS Amplicon Sequencing:
A well-established method for microbial identification and phylogeny studies in complex microbiomes or environments.
16S Amplicon Sequencing:
Specifically used to study bacterial and archaeal diversity and abundance.
Utilises the 16S ribosomal RNA gene, with its sequence variations aiding in species identification.
The sequencing process typically involves PCR amplification of the gene followed by Sanger sequencing or next-generation sequencing.
18S rRNA and ITS Sequencing:
Commonly applied in fungal studies due to the increased hypervariable domains in 18S rRNA.
The Internal Transcribed Spacer (ITS) region is a universal fungal barcode marker, facilitating broad-range fungal identification, such as pathogenic moulds.
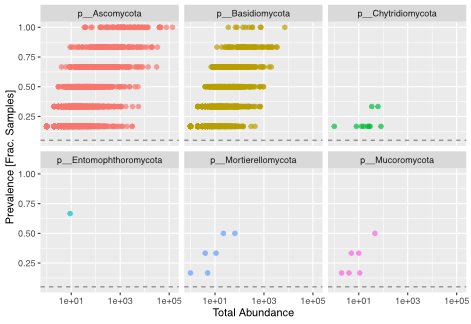
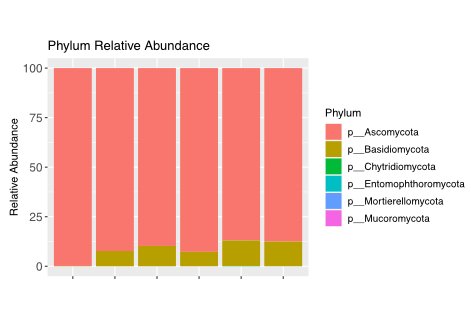
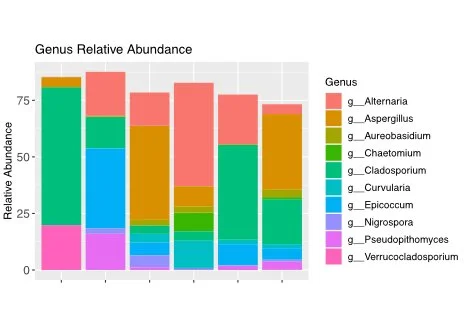
These sequencing methods provide invaluable insights into microbial communities, allowing researchers to identify species and estimate their relative abundance. The choice of sequencing approach depends on the specific goals of the study and the nature of the environmental samples under investigation.